Welcome to the ELEKTRONN documentation!¶
Note
ELEKTRONN 1.0 has been superceded by the more flexible, PyTorch-based elektronn3 library. elektronn3 is actively developed and supported, so we encourage you to use it instead of ELEKTRONN 1.0.
ELEKTRONN is a highly configurable toolkit for training 3D/2D CNNs and general Neural Networks. The package includes a sophisticated training pipeline designed for classification/localisation tasks on 3D/2D images. Additionally, the toolkit offers training routines for tasks on non-image data.
It is written in Python 2 and based on theano, which allows CUDA-enabled GPUs to significantly accelerate the pipeline.
ELEKTRONN was created by Marius Killinger and Gregor Urban at the Max Planck Institute For Medical Research to solve connectomics tasks.
The design goals of ELEKTRONN are twofold:
- We want to provide an entry point for researchers new to machine learning. Users only need to pre-process their data in the right format, and can then use our ready-made pipeline to configure fast CNNs featuring the latest techniques. There is no need for diving into any actual code.
- At the same time we want give people the flexibility to create their own Neural Networks by reusing and customising the building blocks of our implementation. To leverage the full flexibility of ELEKTRONN modifying the pipeline is encouraged.
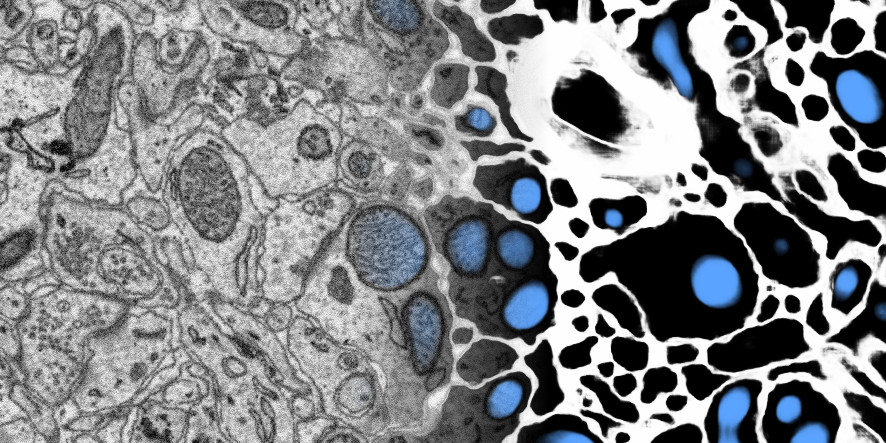
Results example: membrane and mitochondria probability maps. Predicted with a CNN with recursive training. Data: zebra finch area X dataset j0126 by Jörgen Kornfeld.
Main Features¶
- Neural Network:
- 2D & 3D Convolution/Maxpooling layers (anisotropic flters supported)
- Fully-connected Perceptron layers
- Basic recurrent layer
- Auto encoder
- Classification or regression outputs
- Common activation functions (relu, tanh, sigmoid, abs, linear, maxout)
- Dropout
- Max Fragment Pooling for rapid predictions
- Helper function to design valid architectures
- Optimisation:
- SGD+momentum, RPROP, Conjugate Gradient, l-BFGS
- Weight decay (L2-regularisation)
- Relative class weights for classification training
- Generic optimiser API that allows integration of alternative optimisation routines (e.g. from scipy)
- Training Pipeline:
- Fully automated generation of batches
- Data augmentation methods:
- Translation
- Flipping, transposing
- Continuous rotation
- Elastic deformations
- Histogram distortions
- All done online in background processes
- Training with lazy labels (partially labelled data)
- Console interaction during training (allows e.g. to change optimiser meta-parameters)
- Visualisation of training progress (plots and preview predictions)
